Part:BBa_K771002
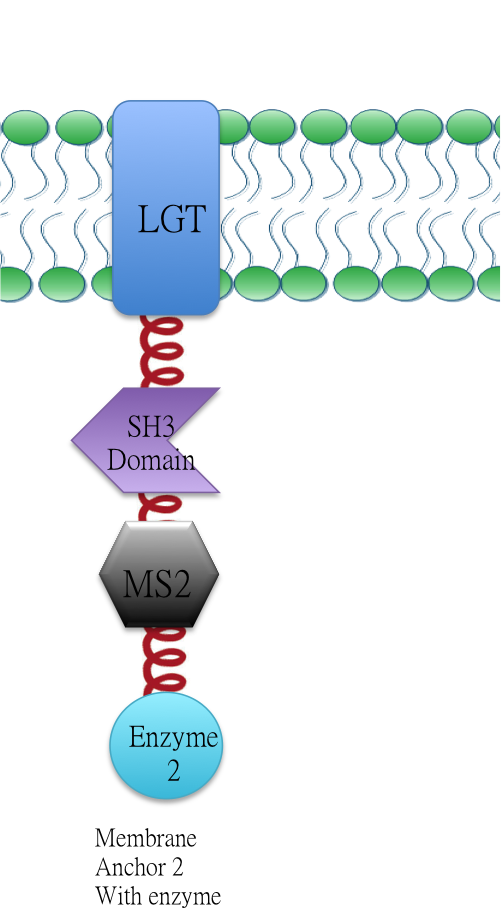
Membrabe Anchor 2:ssDsbA-Lgt-SH3 Domain-MS2
Membrane anchor 2:RNA aptamer binding protein MS2 (BBa_K771112) fused with SH3 domain(BBa_K771107) and membrane protein Lgt(BBa_K771102).
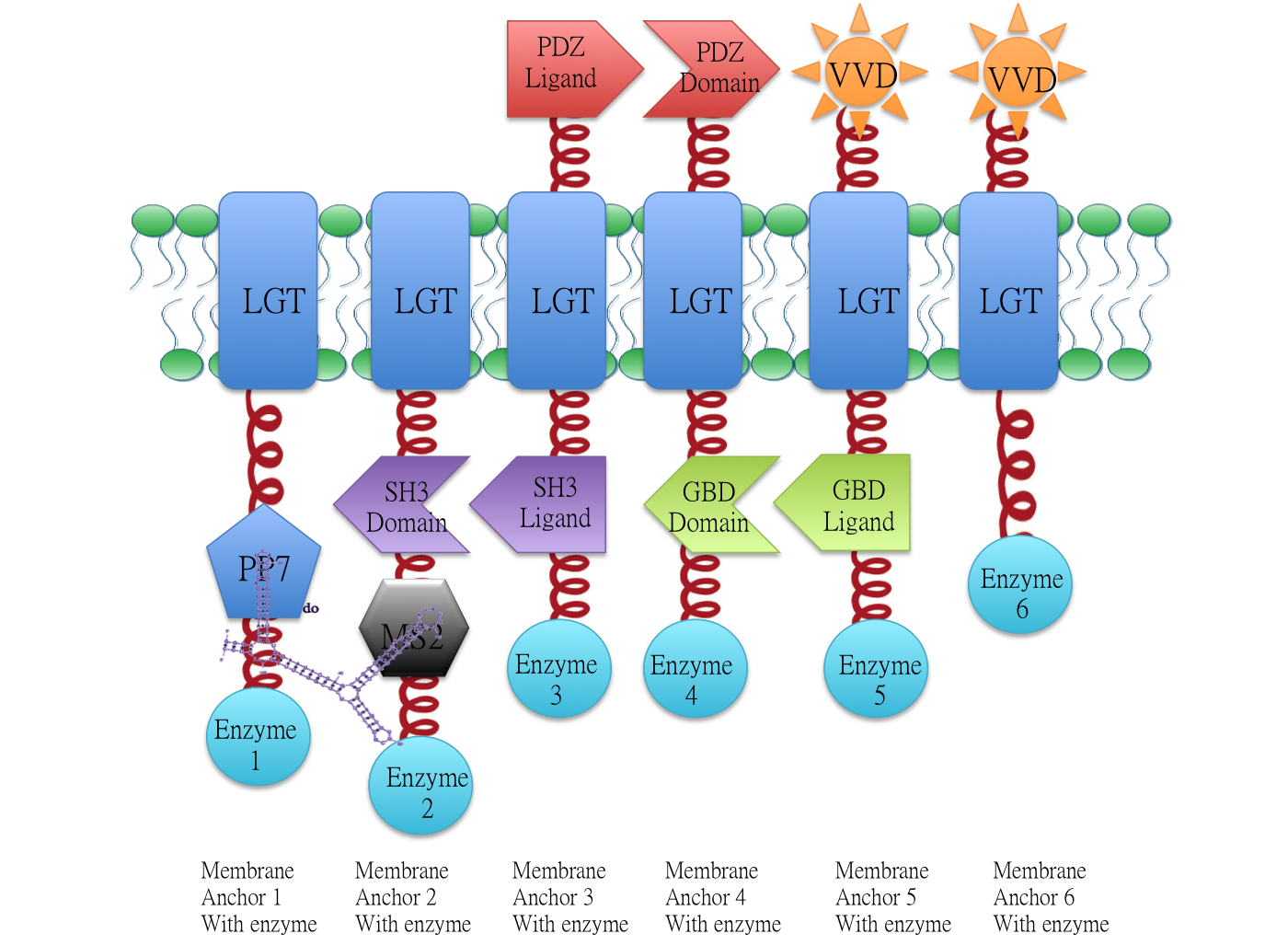
Overview: Membrane Scaffold System
Demonstration of the whole Membrane Scaffold device. Membrane Anchor 2, 3, 4, 5 consitutively aggregate while Membrane Anchor 1 and 6 only aggregate with constitutive cluster (Membrane Anchor 2, 3, 4, 5) when certain signal is present.
Backgroud
Signal Induced Aggregation
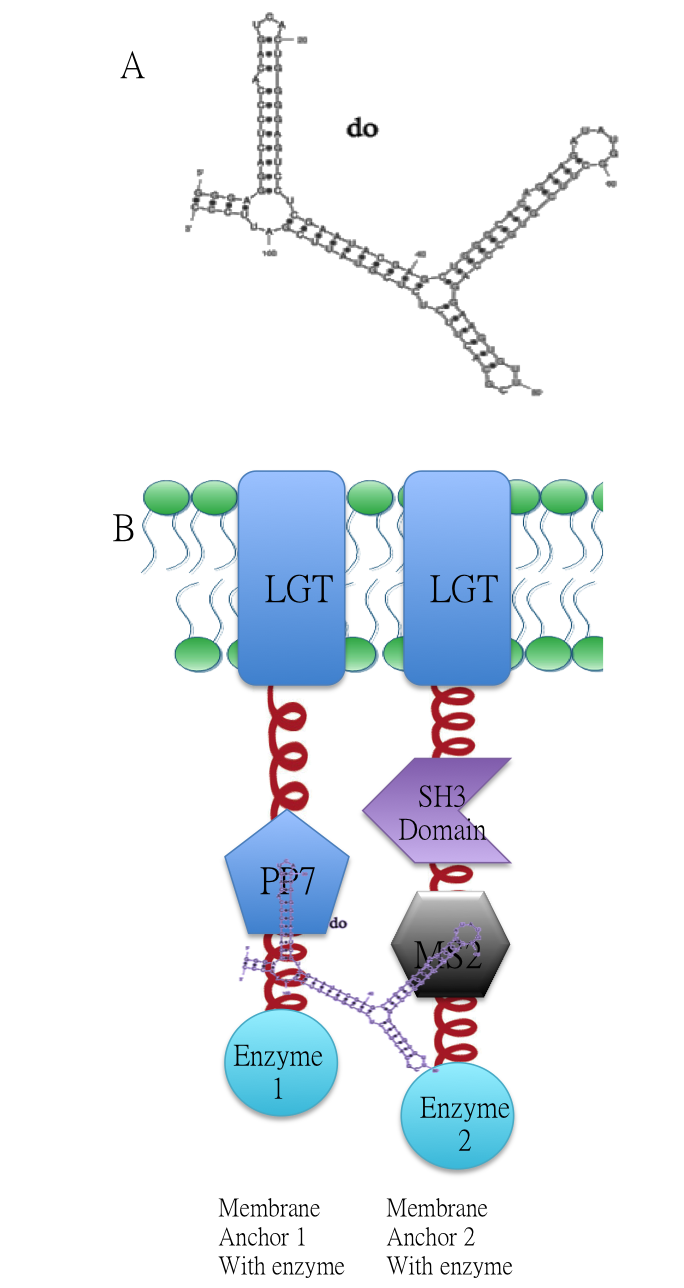
Membrane Anchor 1 and 2 act as a controlling point to determine whether certain pathway (in this case involving enzyme 1, 2, 3, 4 and 5) is activated or not. RNA molecule D0(BBa_K771009) with PP7 and MS2 aptamer domain acts as the signal.
A.Sketch of signal RNA molecule (called D0) which consists of PP7 and MS2 aptamer domains. B: Through fusing PP7 and MS2 RNA biding domain (BBa_K771111 BBa_K771112) to our membrane device, RNA molecule D0(BBa_K771009) in Figure A can function as a bridge to connect different proteins, thus decreasing distance between corresponding enzymes.
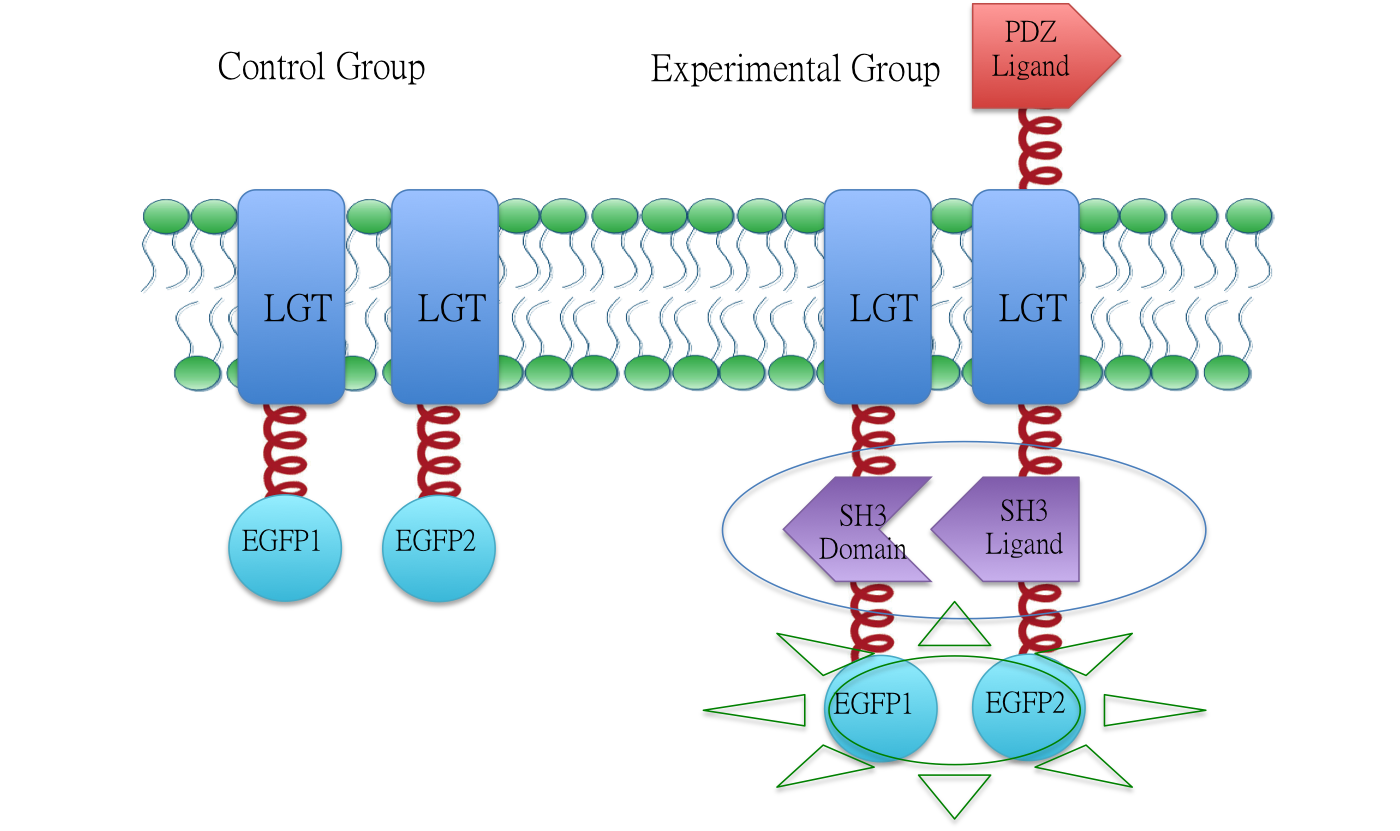
Constitutive Aggregation
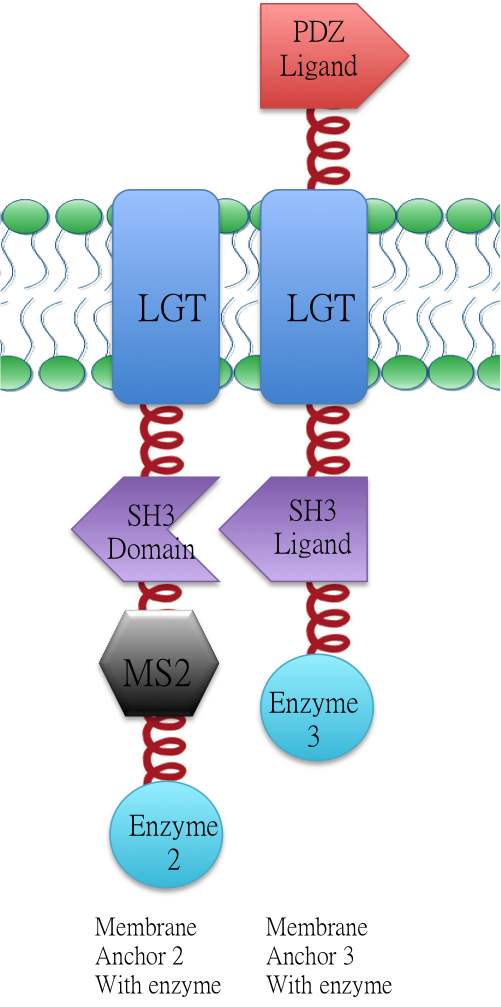
Membrane Anchor 2 and 3 constitutively aggregate through SH3 domain(BBa_K771107) and SH3 ligand (BBa_K771108).
Design
RNA aptamer binding protein MS2(BBa_K771112) fused with SH3 domain(BBa_K771107) and membrane protein Lgt (BBa_K771102). When RNA molecule D0 is present, Membrane Anchor 1(BBa_K771001) is expected to dimerize with Membrane Anchor 2 through MS2(BBa_K771112) and PP7(BBa_K771111) domain.
Characterization
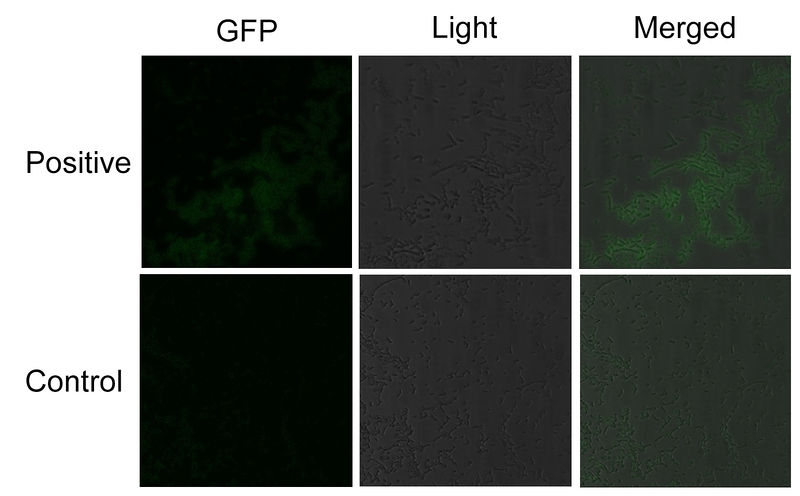
To testify the constitutive aggregation of Membrane Anchor 2, 3 and 4, we conducted Fluorescence Complementation Assay.
In fluorescence complementation assay, proteins that are postulated to interact are fused to unfolded complementary fragments of EGFP and expressed in E.coli. Interaction of these proteins will bring the fluorescent fragments within proximity, allowing the reporter protein to reform in its native three-dimensional structure and emit its fluorescent signal. EGFP was split into two halves, named split EGFP1 (BBa_K771113) and split EGFP 2(BBa_K771114) respectively. If there is interaction between two proteins which were fused with 1EGFP and 2EGFP, it is expected that fluorescence should be observed. Otherwise, no fluorescence could be observed.
Membrane Anchor 2 without MS2 and Membrane Anchor 3
We fused split EGFP 1 and 2 to Membrane Anchor 2 without MS2(BBa_K771007) and Membrane Anchor 3 to test whether Membrane Anchor 2 without MS2 and Membrane Anchor 3 could constitutively aggregate. Proteins in control group are not expected to aggregate like in experimental group.We coexpressed proteins in experimental group and control group respectively in E.coli. After induction for 6 hours, bacteria samples were taken for fluorescence observation.
The result shows that Membrane Anchor 2 without MS2 and Membrane Anchor 3 could constitutively aggregate through SH3 domain and ligand.
Related Biobrick
<br\>Membrabe Anchor 1([BBa_K771001]) <br\>Membrabe Anchor 3([BBa_K771003]) <br\>Membrabe Anchor 4([BBa_K771004]) <br\>Membrabe Anchor 5([BBa_K771005]) <br\>Membrabe Anchor 6([BBa_K771006])
Reference
Delebecque, C. J., A. B. Lindner, et al. (2011). "Organization of intracellular reactions with rationally designed RNA assemblies." Science 333(6041): 470.
Dueber, J. E., G. C. Wu, et al. (2009). "Synthetic protein scaffolds provide modular control over metabolic flux." Nature biotechnology 27(8): 753-759.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 883
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 504
Illegal BsaI site found at 1609
Illegal SapI.rc site found at 1150
| n/a | Membrabe Anchor 2:ssDsbA-Lgt-SH3 Domain-MS2 |